Structure prediction
We retrained AlphaFold-FineTune (AF-FT) on peptide/HLA-I 9mer structures from HLA3DB to create AF-FT-HLA3DB. We find competitive accuracy with significant increases in throughput versus RepPred (30 min vs 3 mins). The repository, with installation and setup instructions, can be found here.
Benchmark results
We benchmarked AF-FT-HLA3DB on the same structures used to benchmark RepPred. We performed a leave-one-out validation where we also removed homologous structures during template selection. Accuracy is competitive across the board, with significant improvement in the A02 BB1 category.
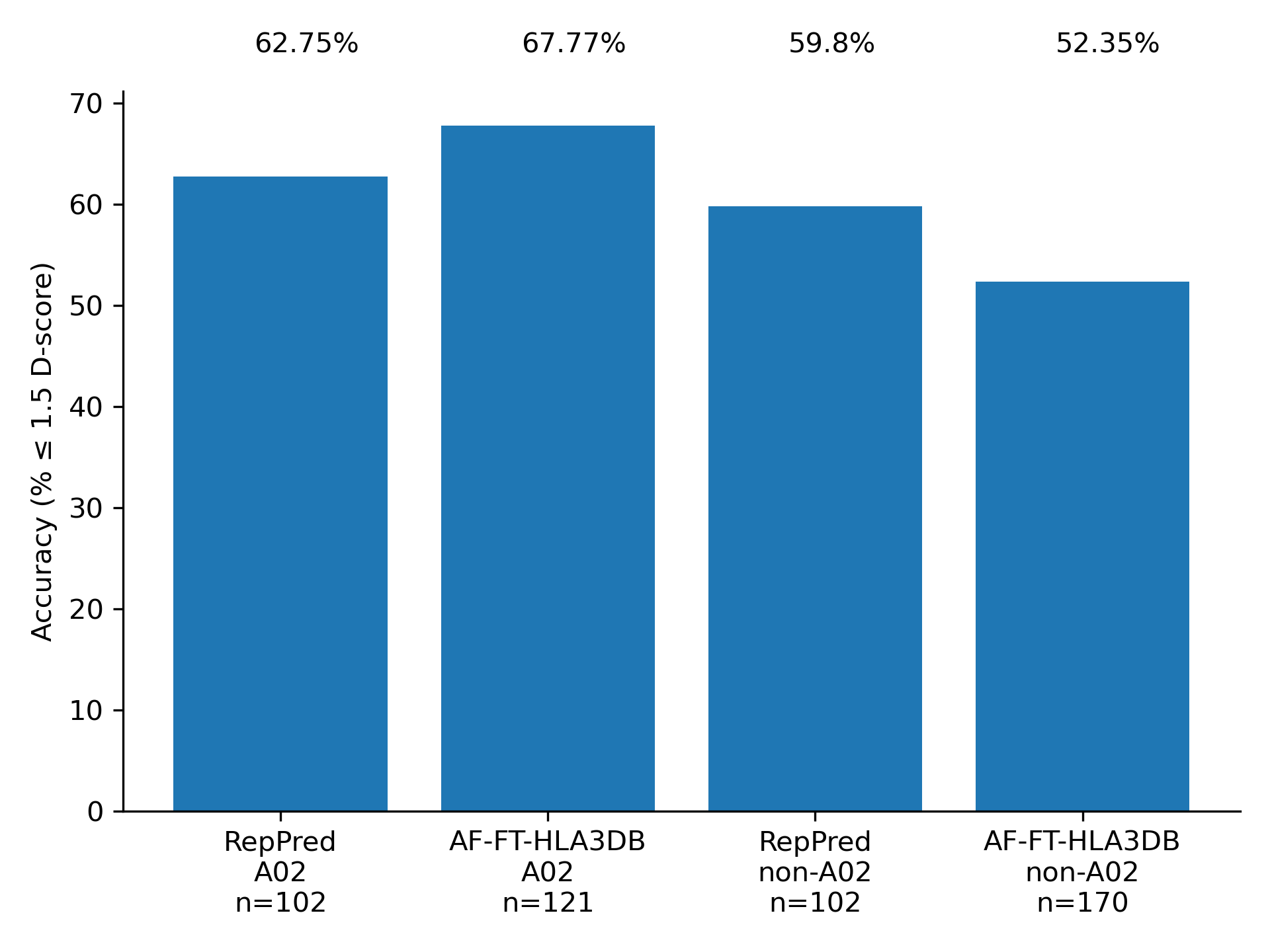
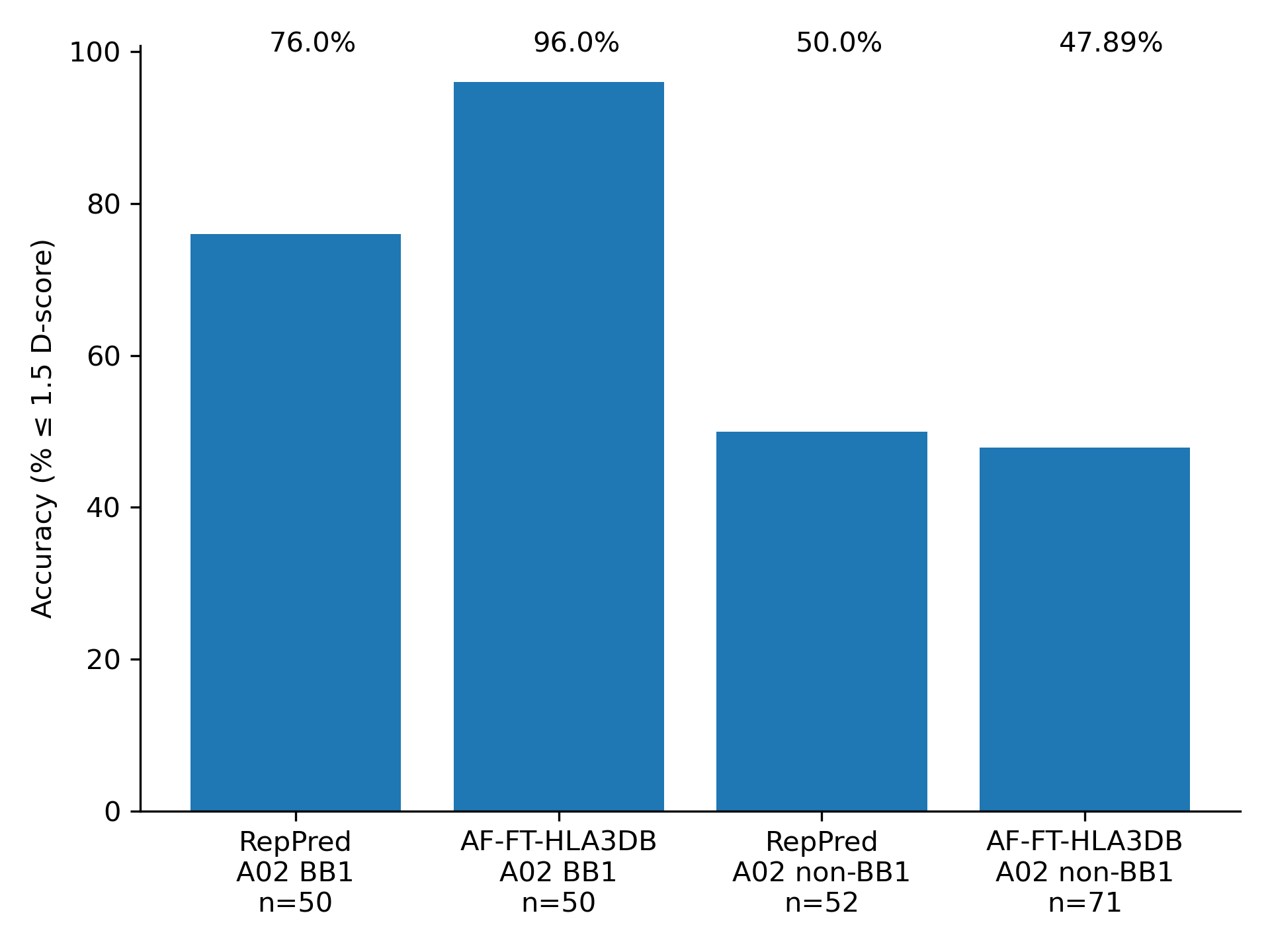
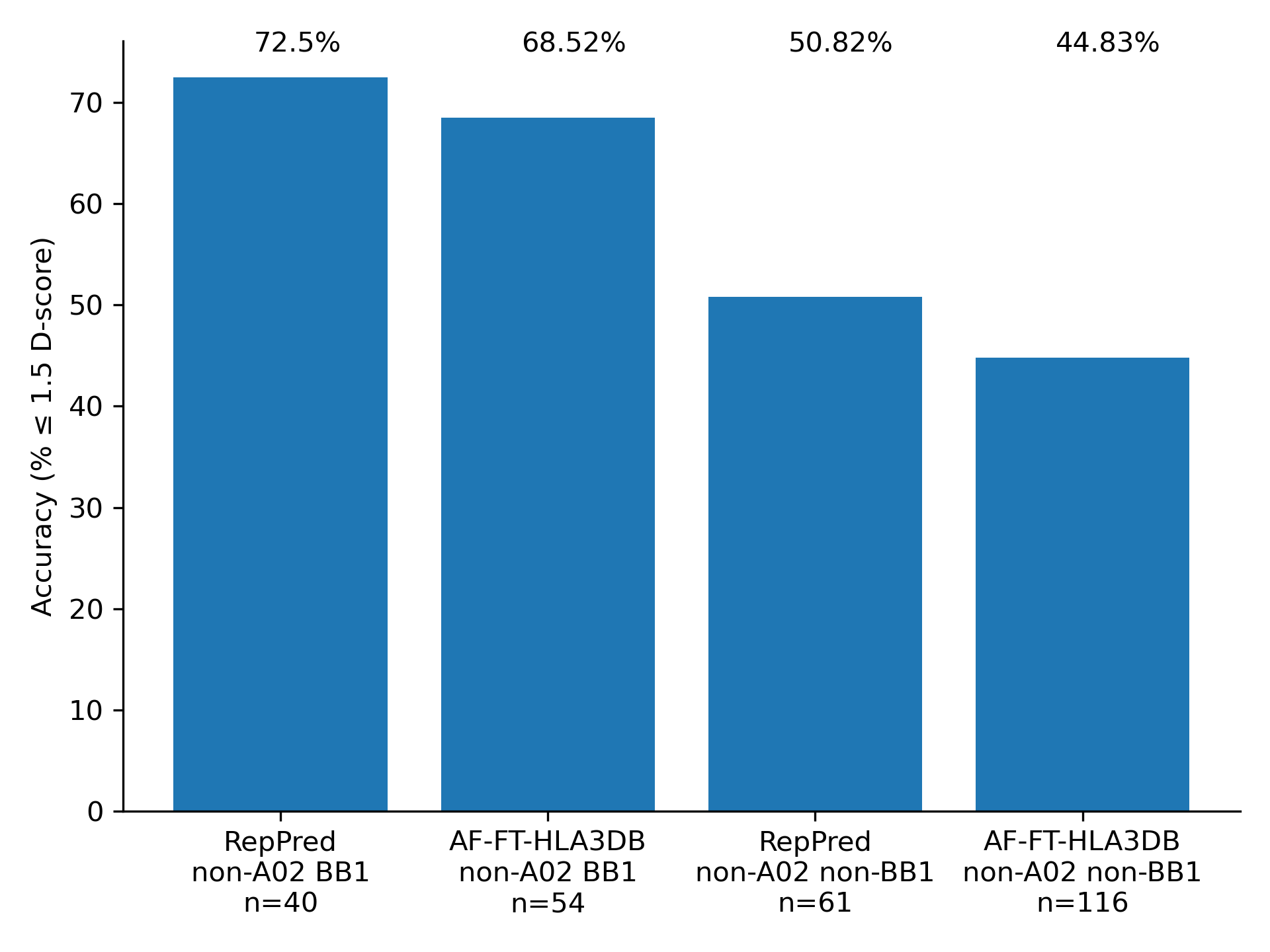
References
Motmaen et al. Peptide-binding specificity prediction using fine-tuned protein structure prediction networks. Proc Natl Acad Sci USA. 2023 21 Feb. doi: 10.1073/pnas.2216697120
Gupta et al. HLA3DB: comprehensive annotation of peptide/HLA complexes enables blind structure prediction of T cell epitopes. Nat Commun 2023 10 Oct. doi: 10.1038/s41467-023-42163-z